Machine Learning in Klustron
Machine Learning in Klustron
Objective:
Every company seeks to improve its decision-making processes with minimal costs through the use of machine learning. For most databases, users need to perform machine learning processes outside the database. However, this is not the case with Klustron, which includes multiple extensions for other languages. Users can train and utilize machine learning algorithms without leaving Klustron.
Running machine learning algorithms in Klustron has several advantages. Firstly, unlike running machine learning algorithms in PostgreSQL, running them in Klustron is not limited by the hardware resources of a single server. It utilizes the computing resources of many servers to work with larger datasets more rapidly. This is due to Klustron's multi-level parallel execution mechanism, which allows it to use the CPU and memory of multiple servers to perform the same machine learning task.
Additionally, compared to running machine learning algorithms outside the database, having a complete infrastructure within the database system to deal with large-scale data issues, including temporary tables, buffer pools, and indexes, is a significant advantage. Running machine learning outside the database either requires manually implementing these infrastructures (which is highly complex) or is limited to operating in memory, severely affected by the operating system frequently paging due to data exceeding memory.
Moreover, running machine learning algorithms inside the database avoids the problems associated with transferring large amounts of data across network nodes, which also helps enhance the performance of machine learning algorithms.
Klustron not only supports running machine learning algorithms through stored procedures calling popular machine learning libraries like PyTorch but will also support common machine learning algorithm extensions like PostgresML and Apache MadLib in version 1.3 to enable more powerful and high-performance machine learning algorithms.
Various machine learning libraries and extensions from the PostgreSQL ecosystem, previously limited by single-machine hardware resources in PostgreSQL, are no longer restricted in Klustron. They can handle larger amounts of data with higher performance and shorter processing times. Additionally, these machine learning algorithms benefit from Klustron's multi-level parallel query processing capabilities, achieving higher performance.
Thus, Klustron significantly amplifies the capabilities of these in-database machine learning components, providing users with robust and high-performance machine learning processing capabilities. Let's take a look at how to use PLPython to execute K-means (one of the most popular unsupervised learning algorithms) directly in Klustron.
01 Import Sample Data
We will use the Iris sample data, which can be obtained from the following link:
https://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data
Place it in the directory /kunlun.
Then, log in to the compute node using the pgsql client to create a user and database:
psql -h 192.168.40.152 -p 47001 postgres
create user kunlun_test with password 'kunlun';
create database testdb owner kunlun_test;
grant all privileges on database testdb to kunlun_test;
alter user kunlun_test with superuser;
\q
User kunlun_test logs into testdb, creates a test table iris, and imports the data:
psql -h 192.168.40.152 -p 47001 -U kunlun_test testdb
create table iris (
sepal_length REAL,
sepal_width REAL,
petal_length REAL,
petal_width REAL,
species varchar(20)
);
\copy iris from '/kunlun/iris.data' delimiter ',';
02 Install Dependencies
2.1 Install Python3
If the system does not have Python3, it will need to be installed:
yum install python3
yum install python3-devel
Verify that Python3 has been successfully installed:
python3 -V
pip3 -V
Set up symbolic links for Python and pip, which allow direct use of python and pip:
ln -s /etc/alternatives/python3 /usr/bin/python
ln -s /etc/alternatives/pip3 /usr/bin/pip
2.2 Install PL/Python Extension
Klustron supports the plpython3 extension out-of-the-box starting from version 1.3.1.
2.3 Install Other Python Packages
su - kunlun
pip install --user scikit-learn
pip install --user pandas
03 Running the Kmeans Algorithm in Klustron
3.1 Create the plpython3 Extension
psql -h 192.168.40.152 -p 47001 -U kunlun_test testdb
CREATE EXTENSION plpython3u;
3.2 Create the Kmeans Function
CREATE OR replace FUNCTION kmeans(input_table text, columns text[], clus_num int) RETURNS bytea AS
$$
from pandas import DataFrame
from sklearn.cluster import KMeans
from pickle import dumps
from pandas import pandas as pd
all_columns = ",".join(columns)
if all_columns == "":
all_columns = "*"
rv = plpy.execute('SELECT %s FROM %s;' % (all_columns, plpy.quote_ident(input_table)))
frame = []
for i in rv:
frame.append(i)
df = DataFrame(frame).apply(pd.to_numeric, errors="ignore")
kmeans = KMeans(n_clusters=clus_num, random_state=0).fit(df._get_numeric_data())
return dumps(kmeans)
$$ LANGUAGE plpython3u;
This function first generates a string from the passed columns, or replaces it with * if an empty array is passed, then builds and executes a query using PL/Python's execute function. After building and executing the query, it converts the results to a DataFrame and transforms the numeric variables into numeric types (by default they might be interpreted as other types). Then, it calls kmeans, passing the number of input groups as a parameter, which represents the number of clusters you want to obtain. Finally, it dumps the model into Pickle and returns the object stored in Pickle. Pickling is necessary for later restoring the model, otherwise Python cannot directly restore the kmeans object from the byte array returned by Klustron.
3.3 Store the Model
After creating the model, it needs to be stored in a table for later use in predictions:
CREATE TABLE models (
id SERIAL PRIMARY KEY,
model BYTEA NOT NULL
);
Call the previously created kmeans function to store the generated Kmeans model in the table:
INSERT INTO models(model) SELECT kmeans('iris', array[]::text[], 3);
3.4 Display Model Information
We can display the model information with select * from models;, but the result is not very readable.
We know all kmeans objects have an attribute cluster_centers_, which is the location of centroids. The centroids are the mean vector of each group, i.e., the average of each variable in each group. Essentially, they are stored as numpy arrays, but since plpython cannot handle numpy arrays, we need to convert them to a list of lists. This is why the returned object is an output listing each row, resulting in a list of lists where each sublist represents a centroid of a group. Obtain the centroids by creating the following function:
CREATE OR replace FUNCTION get_kmeans_centroids(model_table text, model_column text, model_id int)
RETURNS SETOF real[] AS
$$
from pandas import DataFrame
from pickle import loads
rv = plpy.execute('SELECT %s FROM %s WHERE id = %s;' % (plpy.quote_ident(model_column), plpy.quote_ident(model_table), model_id))
model = loads(rv[0][model_column])
ret = map(list, model.cluster_centers_)
return ret
$$ LANGUAGE plpython3u;
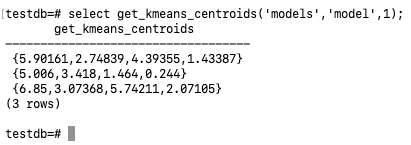
Finally, use the following SQL to obtain the centroids for the 3 clusters:
select get_kmeans_centroids('models','model',1);
3.5 Using the Model for Prediction
The method for prediction is to pass a set of variables and then determine their corresponding group. Create the following prediction function:
CREATE OR replace FUNCTION predict_kmeans(model_table text, model_column text, model_id int,
input_values real[]) RETURNS int[] AS
$$
from pickle import loads
rv = plpy.execute('SELECT %s FROM %s WHERE id = %s;' % (plpy.quote_ident(model_column), plpy.quote_ident(model_table), model_id))
model = loads(rv[0][model_column])
ret = model.predict(input_values)
return ret
$$ LANGUAGE plpython3u;
Pass a set of iris petal and sepal lengths and widths to determine its species:
select predict_kmeans('models','model',1,array[[0.5,0.5,0.5,0.5]]);

Using Kmeans model to predict the variety based on the lengths and widths of sepals and petals.
